Publication summaries
Here I will share short informal discussions of the most recent papers (and maybe also some older ones).
RING dimerisation drives higher-order organisation of SINA/SIAH E3 ubiquitin ligases
Coste, F.✉, Mishra, A., Chapuis, C., Mance, L., Pukało, Z., Bigot, N., Goffinont, S., Gaudon, V., Garnier, N., Talhaoui, I., Castaing, B., Huet, S.✉, Suskiewicz, M.J.✉
RING dimerisation drives higher-order organisation of SINA/SIAH E3 ubiquitin ligases. The FEBS Journal, 5/2/2025.
Link to the article. If the previous link does not work to grant open access, you can access the authors’ version here
Happy to share our new study showing that the Ub E3 ligase SIAH1, which has been known to dimerise via its C-term SBD, also dimerises via its N-term RING. When these tendencies combine in the full-length protein, SIAH1 forms multimers, which might explain its clustering in cells & preference for aggregated/multimeric substrates.
The project started when Zosia was a summer student, & we discussed the great paper by Słabicki et al. The ubiquitylation of BCL6 by SIAH1 is boosted when BCL6 gets multimerised by a small molecule. Why is SIAH1 preferentially targetting the multimeric version of a protein? We reasoned that SIAH1 might also be a multimer & recognise multimers through avidity.
We crystallised and Franck solved the crystal structure of the dimer formed by the RING-ZnF1 part of SIAH1. We confirmed dimerisation in solution with SEC-RALS/LALS, & validated the V90R mutation as blocking RING dimerisation. Aanchal did some lovely AlphaFold modelling showing dimerisation across the SINA/SIAH family. One should add that the SINA/SIAH family is present across many species from plants to humans and is highly conserved. What we say of SIAH1 in this paper, most likely applies to all family members, and we do perform some experiments with drosophila SINA and human SIAH2.
Here’s what happens in cells: mCherry-tagged SIAH1 (admittedly, somewhat overexpressed) localises to elongated clusters, but you need to have both RING and SBD for that, and no V90R mutation, otherwise SIAH1 is mostly diffuse. When we look at the colocalisation with an aggregated substrate, Synphilin-1, SIAH1 WT does colocalise, but this is largely prevented when we block RING domain dimerisation with the V90R mutation. This leads us to a model where multimerisation allows high-avidity binding to multivalent substrates.
Lastly, Lucija performed nice in-vitro ubiquitylation assays showing that the V90R mutant of the RING-ZnF1 fragment is deficient in making long ubiquitin chains. So there is also a role for RING dimerisation in catalysing polyubiquitylation, as seen for other dimeric RING Ub E3s.
Overall, this side project is related to our ongoing interest in how multimerisation of substrates and ligases - particularly filament formation - affects SUMOylation and ubiquitylation processes. I thank all colleagues and collaborators involved in this project and You for your attention ;).
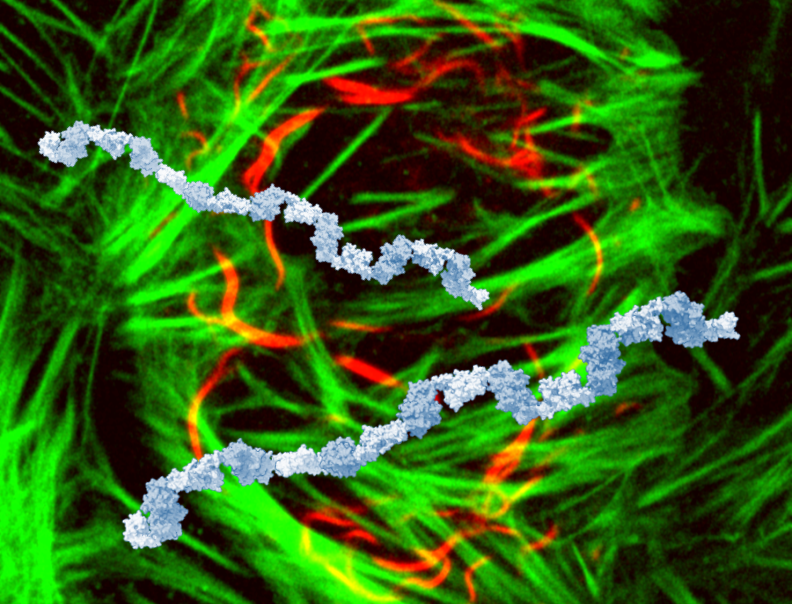
This is a cover design that wasn’t accepted, showing SIAH1 multimers on top of cells (SIAH1 red, actin green). We did go on the FEBS J cover, but with a different image. Of note, we were not able to visualise SIAH1 chains in vitro, as we could only purify N- or C-term halves of SIAH1 separately: the blue chains are models made from combined dimeric structures of each half.
Dynamic BTB-domain filaments promote clustering of ZBTB proteins
Mance, L., Bigot, N., Sánchez, E. Z., Coste, F.✉, Martín-González, N., Zentout, S., Biliškov, M., Pukało, Z., Mishra, A., Chapuis, C., Arteni, A.-A., Lateur, A., Goffinont, S., Gaudon, V., Talhaoui, I., Casuso, I., Beaufour, M., Garnier, N., Artzner, F., Cadene, M., Huet, S., Castaing, B., Suskiewicz, M. J.✉.
Dynamic BTB-domain filaments promote clustering of ZBTB proteins. Molecular Cell, 84(13), 2490-2510. (2024).
Link to the article. If the previous link does not work, you can access the authors’ version here
In this paper, we show that BTB domains of ZBTB proteins incl. ZBTB8A or ZBTB18 make homomultimers, which concentrates them in nuclear foci and contributes to their function as repressors. We demonstrate & characterise this property using crystallography, various biophysical methods, and some cell biology. BTB filaments are built of head-to-head dimers connected tail to tail. They differ in topology from head-to-tail filaments formed by known polymerising domains such as SAM. These differences have implications for evolution & prediction of filaments, as discussed in our paper.
We got interested in ZBTBs, because we work on a PTM called SUMOylation, and they are among the most highly SUMOylated proteins. We thought the BTB domains might interact with the SUMO E2 enzyme, UBC9. Why? In at least one ZBTB protein (ZBTB1), the SUMOylation of this protein was shown to depend on the presence of the BTB domain, even though the BTB domain is physically far away from the SUMOylation site. Also, in SLX4, which was reported to have a SUMO E3 ligase activity, there is a BTB domain that was shown to contributed to that proposed activity. Combined with the fact that ZBTB proteins are very highly SUMOylated and several of them have been reported to interact with UBC9 (ZBTB8A, ZBTB9, ZBTB7A, ZBTB26, ZBTB2, ZBTB16, ZBTB1), it looked like the BTB domain might be a UBC9-interacting domain. We began with the BTB domain of ZBTB8A, which was modelled moderately convincingly to interact with UBC9 by AlphaFold2. Alas, despite these various hints, we did not find any clear experimental evidence that BTB domains of ZBTBs interact with UBC9. Instead, it turned out they make filaments, which we investigated in detail producing this study. We think filamentation which might indirectly promote SUMOylation (we’re now working on this).
Among other things, the fact that we found a filamentation propensity in a protein family that has been considered relatively well characterised suggests that there are more filament-forming proteins than we currently think. One reasons to think so is that filament-forming proteins are frequently insoluble upon recombinant overexpression, and therefore difficult to study. It’s exciting times for this field, because AlphaFold can sometimes predict this tendency, thus facilitating the search for new filament-forming proteins. The tendency to form filaments might explain some instances of foci formation in cells - an alternative, or perhaps complementary, explanation to phase separation.
It was great fun to make this chance discovery and build a project around it. It started as a master & now PhD project of Lucija Mance, a great PhD student. Many thanks to Franck, Sébastien, Martine, and all other great coauthors and collaborators from Orléans, Rennes, Paris/Gif-sur-Yvette, Marseille. And we are also very happy that we could coordinate a back-to-back publication with Paul Park, Jiho Park, Fischer and Ebert groups from Dana-Farber/Harvard. Their paper has just been published as well.
DELTEX E3 ligases ubiquitylate ADP-ribosyl modification on nucleic acids
Kang, Z. ✉, Suskiewicz, M.J., Chatrin, C., Strømland, Ø., Dorsey, B. W., Aucagne, V., Ahel, D. ✉, Ahel, I. ✉
Nucleic Acids Research, 24 November 2023, gkad1119
Link to the open-access article
This is a continuation of a study published last year and available here. Both were spearheaded by Kang and Ivan with contribution from other authors including some from me, especially on the previous paper. I was also very happy that this became a collaboration with another scientist from CBM Orléans, our great chemist friend, Vincent Aucagne.
For those of you who are not familiar with ADP-ribosylation, I should maybe start by saying that NAD+ is not only a central energy molecule in the cell, but it can also be used for a particular type of protein modification called ADP-ribosylation, catalysed by the family of enzymes called PARPs. During ADP-ribosylation a small part of NAD+ called nicotinamide is leaving and the rest of NAD+ (the ADP-ribose part) becomes covalently attached to a protein through the carbon to which nicotinamide was attached.
A few PARPs seem to be pseudoenzymes that are actually inactive. One of those is PARP9, which makes a constitutive complex with a ubiquitin E3 ligase DTX3L. I said that PARP9 seems inactive, but when it is together with DTX3L and when ubiquitin cascade components are present (an E1 enzyme, an E2 enzyme, ATP), a robust reaction between NAD+ and ubiquitin is observed, as demonstrated by the Paschal group six years ago.
In the presence of these components (PARP9, DTX3L, E1, E2, and ATP), NAD+ (or a part of it) becomes somehow joined with ubiquitin. So what is this reaction? It was proposed to be ADP-ribosylation of ubiquitin: nicotinamide presumably leaves from NAD+, the ADP-ribose part of NAD+ becomes attached to ubiquitin through the carbon atom on ADP-ribose to which nicotinamide was attached. Just like in any other protein ADP-ribosylation event catalysed by PARPs.
I should say that a few years later the Huang group - and especially Chatrin Chatrin, who has since joined Ivan’s group and is a co-author on Kang’s NAR paper - showed that, in fact, PARP9 is dispensible for the reaction. You do need a DTX protein - either DTX3L or one of its human homologous from the DTX family - as well as E1, E2, and ATP. A minimal fragment of DTX3L that you need is the RING domain and an adjecent NAD+/ADP-ribose binding domain called DTC.
So what did Kang’s two papers bring to this story? They showed - to my mind conclusively - that this mysterious in vitro reaction between NAD+ and ubiquitin that DTX proteins catalyse is not canonical ADP-ribosylation. Nicotinamide is not leaving, NAD+ stays intact, and ubiquitin becomes covalently linked through an esther bond to one of the ribose hydroxyl groups of NAD+. Vincent Aucagne (with some assistance from Hervé Meudal from our NMR platform) has been instrumental in identifying where Ub becomes attached.
You do not displace nicotinamide during the reaction, and, in fact, nicotinamide does not need to be present at all. Thus, the DTX reaction actually also works with ADP-ribose. In fact, ADP-ribose is preferred over NAD+ as a substrate. And - most interestingly - DTX enzymes can ubiquitylate ADP-ribose that is attached to a protein or a peptide through a prior ADP-ribosylation reaction.
This is new chemistry, and potentially very cool one, because you ubiquitylate a protein not on a lysine residue, which would be typical, but on an ADP-ribose post-translational modification. And as a result you get a dual ubiquitin-ADP-ribose modification that could perhaps have its own distinct function in the cell?
And now a further twist. DTX3L turns out to have nucleic acid-binding domains. It has recently been shown that some PARPs can ADP-ribosylate not only proteins but also nucleic acids (whether that actually happens in the cell is not clear yet). So could DTX3L ubiquitylate ADP-ribose that is attached to nucleic acids? It turns out it can, and it does so more robustly if a full-length protein with nucleic acid-binding domains is used.
An important point: these studies describe fairly robust and specific in vitro reactions, but they remain to be demonstrated in the cell.
And the last point. I am aware that in schematics in both papers there is a mistake in the formula of ADP-ribose: one ribose has wrong stereochemistry. The error originates with me and is embarassing considering I have worked on ADP-ribose for a few years now. I am grateful to a chemist in the room when I gave a talk last summer who pointed out to me something I had always overlooked. I apologise to Kang and other colleagues who, by trusting me, have copied or overlooked this error. I hope this will not distruct the reader from Kang’s elegant experiments and their fascinating conclusions.
ADP-ribosylation from molecular mechanisms to therapeutic implications
Suskiewicz, M. J., Prokhorova, E., Rack, J.G.M., Ahel, I.✉
Cell, 2023, Oct 12;186(21):4475-95.
Link to the open-access article
A new comprehensive review on ADP-ribosylation written together with former colleagues from the Ivan Ahel group at the Dunn School in Oxford (I’m very grateful to Ivan for this opportunity and Evgeniia and Johannes for the work together).
ADP-ribosylation is a fundamental biochemical modification reaction where ADP-ribose is transferred from NAD+ to a substrate (typically a protein). Multiple rounds of ADP-ribosylation can result in the formation of poly(ADP-ribose) chains. ADP-ribose modification can regulate various aspects of biomolecular function, particularly interactions. As there are several protein domains and motifs that recognise ADP-ribosylation, the modification can induce new protein:protein interactions. Inhibitors of the main human ADP-ribosylation enzyme, PARP1, have been successfully used in the clinics to target specific cancer types. In this review, written over the last several months, we attempted to cover a large ground, spanning chemistry, structural biology, enzymatic mechanisms, various cellular pathways, and finally clinical applications.
I am happy with this review except for the positive charge (plus) sign that was put by accident at the production stage on detached nicotinamide in the first figure.
Structural insights into the regulation of the human E2∼SUMO conjugate through analysis of its stable mimetic
Goffinont, S., Coste, F., Prieu-Serandon, P., Mance, L., Gaudon, V., Garnier, N., Castaing, B., Suskiewicz, M. J. ✉
Journal of Biological Chemistry, 2023, 299(7)
Link to the open-access article
During ubiquitylation and related reactions, ubiquitin or a related modifier is first loaded on a Cys residue in an E2 enzyme, producing an E2-modifier thioester. The modifier is discharged from there onto a Lys residue in a protein substrate, often with the help of an E3 ligase.
SUMOylation is an essential ubiquitin-like modification. While ubiquitylation relies on numerous E2s, SUMOylation depends on a single one, UBC9, which is therefore the central protein of the pathway. UBC9’s highly conserved, with sequence changing little from yeast to humans.
Since the E2-modifier thioesters are unstable, our goal here was to produce a stable mimetic of human UBC9-SUMO. We used a strategy developed by the great Lima group for the yeast Ubc9, which involves introducing a Lys residue close in space to the active-site Cys93 of UBC9. We describe our attempt at this strategy in detail, and show that SUMO efficiently moves from Cys93 to a Lys placed in the position 129 through an Ala129Lys mutation, forming a stable molecule with SUMO attached 3 Å away from its native position in the UBC9-SUMO thioester.
We crystallised this mimetic. In the absence of an E3 ligase, it adopts the so-called open conformation, which would likely correspond to an inactive state of the thioester. Indeed, all previous structures of UBC9-SUMO were with an E3 ligase, which stabilised the closed state. Interestingly, in the crystal, the mimetic forms chains via a noncovalent interaction between SUMO from one UBC9-SUMO molectule and UBC9 from the next one. We think such interactions can sometimes form in cells, sterically discouraging the active, closed state of UBC9-SUMO. Another interesting point concerns Cys138, a surface-exposed Cys in UBC9 with unclear function. In our open-conformation crystal structure, Cys138 is close to Cys52 of SUMO and they apparently became crosslinked by DTT. Could these Cys residues form a disulphide bridge under some conditions in cells?
There’re some other elements to the story, too. We’ve put emphasis on detailed description and discussion. We’d be happy if it’s useful to the field.
It’s part of a larger line of research where we’re trying different biochemical and chemical ways of stabilising SUMOylation complexes. It’s the first publication from our SUMOwriteNread project. Our engineer Stéphane Goffinont played the first fiddle (thanks and congratulations!), with assistance from Franck Coste (crystal structure), Pierre Prieu-Serandon, and the rest of the team.
We have a short popular description of the article in English on our Centre’s website.
Updated protein domain annotation of the PARP protein family sheds new light on biological function
Suskiewicz, M. J. ✉✱, Munnur, D.✱, Strømland, Ø.✱, Yang, J. C., Easton, L. E., Chatrin, C., Zhu, K., Baretić, D., Goffinont, S., Schuller, M., Wu, W.-F., Elkins, J. M., Ahel, A., Sanyal, S., Neuhaus, D., Ahel, I. ✉
Nucleic Acids Research, 2023, 51(15):8217-8236
Link to the open-access article
In this side-project paper, we carefully analysed AlphaFold2 models of human members of the PARP protein family, made a comprehensive domain annotation, made new insights into structure & function, and experimentally validated some of them.
We hope that the domain annotation will be a useful resource. We believe all structured domains within PARPs are now labelled. Some have not been reported before - for example, the KH domains, which are potential sequence-specific RNA- or ssDNA-binding domains, within PARP9, 10, & 14.
I did most of the computational analysis and writing, while Deeksha & Øyvind from the Ivan Ahel lab (my former group) have done experiments demonstrating that PARP14 fragments can indeed bind to - and ADP-ribosylate - nucleic acids in vitro. David Neuhaus’ team at MRC LMB Cambridge has contributed an interesting NMR experiment showing that while individual PARP1 domains have independent mobility in a DNA-free state, most of them cluster together upon DNA break binding - except for the BRCT domain, which remains flexible.
In the introduction, we tried to explain in a simple way the principles behind AlphaFold2 and introduce various recent easy-to-use tools for protein analysis. Hopefully this part might be interesting even for people working on some other protein families.
The Institute of Chemistry of the CNRS has dedicated to this paper a nice news & views article in French, and we have a short note in English on our Centre’s website, too.