Blog
Chances are that this blog will die at some point. But it’s a risk I’m willing to take. I will write here short notes, mostly, I guess, on scientific articles I’ve read.
Before a talk (20/06/2025)
Waiting in the park outside, watching people jogging, exercising. A young girl boxer is practicing with her coach, they are punching each other with boxing gloves. I’ve just passed a house with an inscription “Simone Weil philosophe a habité cette maison de 1929 a 1940” and tried talking photo of it with the zoom camera of my laptop, as I don’ have a smartphone, to send it to a friend who is very fond of her.
The second day of the annual SFBBM meeting in Paris won’t start until 8.30 am (it’s 7.20 am now), but I came from Orleans with the first train of the day just to make sure I’m not late as I’m the first one to give a talk today. The first day yesterday was very nice - a wide range of topics and good quality of presentations and posters, some familiar faces whom its a pleasure to see again and talk to. The meeting is held in the pharmacy department, one of the late 19th century’s “churches of science” meant to convey the spirit of positivism and scientism, complete with frescos and sculptures. The conference room itself is not in the best condition, the benches are a bit worn and the walls dusty, but that’s part of the charm of the meeting - that it is relatively modest, using public spaces used normally by student and therefore cheaper to organise. Behind the building there is a lovely small botanical garden.
I have a talk at 8.30 am and whenever I do, I sleep badly. I was woken up by my daughter at 3 am and didn’t fall asleep. Practiced my talk a bit, went to the train station on foot, took the 5-o’clock train, arrived just after 6 am, and then wandered around Paris for an hour murmuring the talk to myself. I made a new introduction - tried to make it a bit interactive - let’s see if it works out or not. I will speak about protein filaments made by ZBTB proteins and SIAH E3 ligases studied by us recently. I will ask people in the room to imagine they are proteins: if they can only use one hand to connect with each other, they can only make pairs, but if they can use both hands, depending on the angle between their two arms, they will make circles or chains. Et cetera. I will not ask people to actually do this but only imagine. It may be a complete fiasco, let’s see.
Will try to find some tea before it starts.
Springtime in the lab (14/04/2025)
Last week was beautifully sunny and warm. Spring has truly arrived, bringing new hope and joy. This week was supposed to be colder and rainier, and I guess it will be, but as I’m writing this, the spring sunrays are squeezing through holes in the roller blind. Hope and joy do not give in easily.
I’m alone for a few days - my family is at my parents-in-law - and my plan is to work a little bit more and finalise one manuscript. We have recently finalised another, smaller one. This year is a bit busy with things we need to finalise. But that’s good, too.
It’s my fourth year in France and in my permanent researcher job, so things don’t strike me as new all the time any more, and there is some sense of recurring yearly cycles, and it has a lot to do with what kind of interns we are having in the team. The spring, for example, is the time when the students from a vocational college (BTS) have just finished their internships, and the 1st year master students join for a two-month project, and the 2nd year master students are more settled into their work and have to start thinking of the writing. I’ve worked closely with one BTS student, one 1st year, and one 2nd year master student this year, and I’ve enjoyed it very much, even if it is sometimes a bit difficult to reconcile doing experiments with a student with other duties.
To continue with good things, our project about filament formation among transcription factors was selected for an HFSP Early Career award, meaning it will be funded. It was a second attempt, after failing to get the funding last year. Failing and having to reapply is never pleasant but it did the project some good - we have matured our ideas and made the project a bit more streamlined. Let’s see what we find.
Review on molecular principles of condensate formation (05/02/2025)
So there we are in Grenoble again, this time first on cryoEM data collection (today and tomorrow), and then on old good X-ray diffraction data collection (tomorrow and the day after).
Sitting and waiting for the data to be collected I have finally finished reading this review by Alex Holehouse and Simon Alberti. I liked it a lot: it is balanced, careful about overstatements or ambiguities, but also bold in making proposals as to how these things called condensates work, or how we might understand them, in an approximated but useful way, to work.
I liked the initial historical bit, bringing from oblivion old papers about ‘quinary protein structure’, and even more so the rest of the ‘Introduction’, where the authors defined the key concepts and distinctions. The biomolecular condensates emerge as molecular networks driven by the ability of some biomolecules to simultanously form more than two (so at least three) homo- or heterotypic interactions (‘multivalency’). With monovalency you get dimers, with bivalency linear filaments, and with at least three interactions you get something that can start forming networks in 2D or 3D. ‘Multivalency’ is a key term here. Thus the controversial ‘phase separation’ just is partitioning due to network formation (so shouldn’t be that controversial after all, I guess, even to classically-inclided biochemists). The authors suggest it might be better to drop the common ‘liquid-liquid’ descriptor, as it is hard to formally demonstrate liquidity as opposed to ‘viscoelasticity’, i.e. some liquid and some solid features. I love how they give their due to all interaction types: from well-defined site-specific ones (between folded domains or a domain & a motif) to ‘chance’ interactions any exposed protein bits - in folded domain or IDRs - can form and which may be significant (particularly when things get locally concentrated), functionally relevant, and evolutionarily selected and maintained. To describe these latter interactions, they use the term ‘chemically specific’ (not sure I’m fully convinced), emphasising that they’re not encoded in specific folds or motifs but in bulk amino acid composition or permissive patterns.
As for the different types of site-specific interactions, they do mention multimerisation of folded domains, including filament formation, a topic of great interest to our team. And they cleverly comment that since the bivalency observed in filaments is not enough to make a network in 2D or 3D, filament-forming proteins will be expected to have additional binding properties as well.
One important fragment: “One common misconception is that IDRs are inherently predisposed to drive phase separation. This is no more the case than the idea that folded domains are inherently predisposed to mediate protein:protein interactions. In both cases, the key feature is the tendency of the proteins to undergo attractive intermolecular interactions, which is governed by the specific amino acid sequence”.
And another important quote: “While it is sometimes suggested that phase separation requires ‘promiscuous’ or ‘non-specific’ interactions, this is a misconception. While promiscuous multivalent interactions can drive phase separatio, so too can highly specific multivalent interactions”. This last quote is very balanced, but the authors do allow hierarchy in condensate formation, with certain proteins acting as ‘scaffolds’ (perhaps more often through site-specific interactions with a dash homomultimerisation?), others as ‘clients’ that join - and necessarily also modify - the crowd, because what counts for final partitioning are all molecules, including ions etc. The focus on the review is on proteins, but they do discuss nucleic acids as well, and even poly(ADP-ribose), a nucleic acid-like polymer dear to my heart. The authors also caution against reading too much into in-vitro phase separation, as “almost any protein will - under an ‘appropriate’ set of solution cond. - be able to undergo self-assembly”. In vitro methods seem best adapted to studying robust site-specific interactions (which may contribute to condensate formation), but condensates themselves should be studied in cells, ideally quantitatively.
Lastly, it was just a pleasure to read. Alex and I did our undergrad together, especially in the first two years, sitting side by side in all tutorials, writing essays on the same topics. And now this - and all his phenomenal work on IDRs etc.! What a privilege to look back on those days!
Two condensate papers (17/01/2025)
The cell is not homogenous and, even disregarding the different specific organelles, it has areas of higher or lower concentration of various components, with many denser bodies or foci composed of proteins and other macromolecules. Thus, many cellular components are not evenly distributed but instead concentrate in distinct bodies/foci, despite these not being delimited by a membrane. We speak of the propensity to cluster or condense; we speak of condensates. And different cellular condensates vary in their composition - meaning there is specificity to the process.
I’m mostly interested in proteins, and more specifically, coming from structural biology, I tend to focus on folded protein domains and interactions in which at least one of the two partners is such a domain (the other can also be a folded domain, or a short peptidic motif that folds upon binding, or a non-protein molecule, e.g. a nucleic acid). Do such canonical interactions explain condensate formation? There are many examples where it looks like they do.
For example, we ourselves have worked (and continue to work) on protein domains that have an ability to homomultimerise in a chain-like manner, and this propensity is required for concentrating a particular class of proteins (e.g. transcription factors called ZBTB) into foci in cells.
I read two papers on condensate formation today, and they made me think of the various interactions that contribute to condensate formation, maturation, and dynamics. The first paper, by the Anthony Leung lab, is consistent with an important role for folded protein domains in condensate formation. The paper is about stress granules, cellular condensates that form under stress and are composed of various proteins, RNA, and a less common biopolymer called poly(ADP-ribose). It shows that a protein called PARP13 affects stress granule size and dynamics, and apparently does so thanks to its multivalent nature, with at least three different types of possible interactions, each mediated by a folded interface within PARP13. PARP13 thus homodimerises and can interact both with poly(ADP-ribose) – which seems key – and with RNA. As a consequence of its multivalency, PARP13 can act as a ‘crosslinker’ that provides vital connections within the noncovalent network that builds stress granules. These connections are not essential for the initial formation of stress granules - other interactions likely play a more fundamental role in maintaining the network - but they do influence the properties of granules, including their fusion and dynamics. This adds some nuance, by the way, to a simple distinction between the formation of a condensate or lack thereof.
The second paper I read today goes a bit against my instinctive focus on folded protein domains, but I don’t think it necessarily goes all the way to support the idea – sometimes entertained – that biomolecular condensation occurs primarily through some sort of nonspecific contacts between disordered regions. This paper is about the transcription factor FOXM1 and condensates it forms in cancer cells. It is true that the key FOXM1 region turns out to be not a folded domain but - by any standard of prediction - a disordered fragment. However, the core of the disordered region in question is not your typical disordered region: it is very highly conserved in sequence, can specifically self-associate with submicromolar affinity, and assembles fibre-like structures in negative-stain EM in vitro. I guess it is some sort of amyloid-like structure rich in beta-strands but more dynamic than conventional amyloids - if that is feasible? I tried AlphaFold and it does predict some sort of amyloid (but then AF almost always does when you ask it to multimerise a short peptide!). In any case, there is no canonical folded domain in sight, but the underlying self-association still likely is structurally defined, as the strict sequence conservation would be difficult to explain otherwise. By the way, the authors of the paper designed a peptide that mimics the key region and which, by interacting with FOXM1, interferes with condensate formation, with beneficial effects in cells and in vivo.
Thus, with these two papers, we are still in the realm of either folded domains or defined, conserved self-interacting motifs. I guess you could have condensates driven by less well-defined (‘fuzzy’) interactions - but you would still need these interactions to be somehow specific, if they really drive specific condensates. And in the protein world, specificity usually, sooner or later, boils down to specificity for particular amino-acid residues (or classes of amino-acid residues) in particular positions, even if these do not form a contiguous motif but are distributed on a surface of a folded domain or as a specific pattern or ‘grammar’ within a disordered region. I do think there might eventually also be a role for less specific, very transient interactions mediated by disordered regions not conserved in sequence - but intuitively I would see such interactions as auxiliary, happening between partners that are already brought into proximity through other means.
I should finish by saying that these are just some loose musings prompted by reading and by extrapolating a structural biologist’s thinking to cell biol… I know there are actual experts working on these things. A friend, Alex Holehouse, has just co-authored a review that seems to cover these topics and which I will try to read next.
Nonscientific reading 2024 (06/01/2025)
In 2024, I kept, for the first time, I think, a clear record of all booked I read, in a chronological order. For many years now, I have a diary, where I write quite a lot every week and I also write on books, but now I added a list where I put down the titles I’ve read at the back. It’s hard to compare with previous years, but it seems like 2024 was a decent reading year for me — at least compared to other years since starting a family. I don’t have much time to read nonscientific things these days, and most of it happens on the tram during my commute back from work or a bit before bed. I try to keep notes on my reading in my diary, which takes some extra time. For this list, I’ve included not only things I read by myself but also a few longer children’s books I’ve read with my daughters (two by Kästner and one by a forgotten Polish author Buyno-Arctowa; we’re currently reading another Kästner). The books are a mix of Polish and English (some of them translated into one of these two languages from another). There wasn’t a specific theme or method to my choices — reading has become a “sphere of freedom” for me, where I try to just follow my intuition or chance. That said, I also try to read books that are gifted or recommended to me.
One book that stood out for me this year was a biography of the film director Éric Rohmer. It allowed me to dive into an art form I know little about and explore one creator’s life and work. My wife and I also watched some of his films available at our local library. I’ve written about it in one of the posts below. I’m glad I tried Trollope this year, an author I hadn’t read before; I’d like to read some more. I also enjoyed the first volume of Stefan Swieżawski’s memoirs (he was a historian of philosophy from Poland). There are two more volumes to go, which I hope to finish in 2025. Some books were short (which can be motivating when keeping a reading list!), but two were longer — the one on Rohmer and the one on Péguy.
Does nonscientific reading have any impact on how we approach science? I like to think it does. At the very least, it helps with writing. But I believe it can do more: it can shape the way we think, structure arguments, and imagine models and metaphors. It can also teach us to observe more keenly, nurture curiosity, and even offer lessons in morality — and I do think morality it one of the key things in science. One of the books I read this year was an essay by Christopher Dawson on the beginnings of the Oxford Movement. For those unfamiliar (it is a bit of a niche topic, I guess), the Oxford Movement was an unusual episode in the history of Anglican theology, marked by going back on some of the precepts of Reformation. Dawson highlights that the movement’s courageous pioneers (well, one of them in any case) were/was inspired by the medieval ideal of chivalry — understood as a selfless and brave dedication to a chosen goal and moral integrity, even in the face of difficulty and criticism. This ideal could be inspiring for scientists as well, couldn’t it? I think there are interesting lessons for scientists in Rohmer and Péguy, and Naipaul is a good teacher of concise style. And Trollope’s portrayal of a career in politics — where ambition meets morality — has many parallels to the world of science.
Nonscientific books read in 2024:
Éric Rohmer: A Biography, Antoine de Baecque & Noël Herpe (English, trans. from French)
Tajemnica Frontenaków, François Mauriac (Polish, trans. from French)
Anatolin, Hans-Ulrich Treichel (Polish, trans. from German)
Uległość, Michel Houellebecq (Polish, trans. from French)
Emil i detektywi, Erich Kästner (Polish, trans. from German)
Miguel Street, V. S. Naipaul (English)
Fantomy, Maria Kuncewiczowa (Polish)
The Spirit of the Oxford Movement, Christopher Dawson (English)
Carnal Spirit: The Revolutions of Charles Péguy, Matthew W. Maguire (English)
The first three volumes of Tintin (French)
Mania czy Ania, Erich Kästner (Polish, trans. from German)
Wielki przełom: 1907–1945, Stefan Swieżawski (Polish)
Kocia mama, Maria Buyno-Arctowa (Polish)
Nie mówię żegnaj, Han Kang (Polish, trans. from Korean)
The Autobiography, Anthony Trollope (English)
Can You Forgive Her?, Anthony Trollope (English)
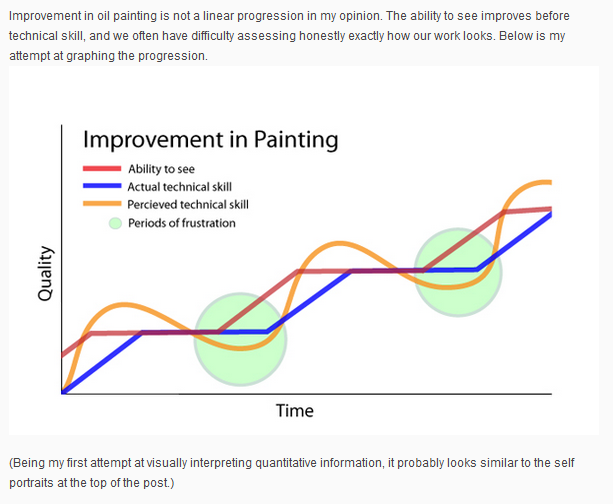
Seeing before knowing how to do (11/12/2024)
I have stumbled upon this graph, by a modern realistic painter Marc Dalessio, from his website. It is about a situation where your ability to paint better has not yet developed, but you can already see more than before and, therefore, are more critical of your current work. According to him, such moments of increased self-criticism generally precede moments of improvement - for him as a painter, at any rate. If seeing differently is actually required to paint differently - but is not by itself sufficient and requires the hand to learn to follow the eye, so to speak - this seems to make sense. I thought that it might apply to scientists as well, offering some comfort to those who are plagued by frequent bouts of self-criticism, as many scientists probably are. Being self-critical assumes we already sense something but do not yet know how to address it - perhaps we sense our writing is not good enough, or our way of asking questions, or a simplistic model we assume, or a simplistic methodology we use. It is comforting to think that such moments might generally precede improved ability to match the increased expectations. The problem is - it never stops…
November update (23/11/2024)
The last time I wrote anything here was just before the EMBO ubiquitin conference held in Cavtat, Croatia. The conference marked an important moment in my life, as it was the first larger conference at which I presented the work done by my colleagues and myself since I started my permanent position in France. Also, and perhaps more importantly, it was the first confernece to which several of us - members of my current research group - went together. Going together as a team to a conference is always special, and when you are partially responsible for the scientific direction of the team, it is extra stressful but also more of an adventure. We shared two little apartments we rented, and the walk to and from the conference venue. We sat next to each other in most talks, then discussed what we heard. The place was beautiful, the atmosphere - welcoming and exciting. I saw some people I’d seen before, others that I’d only heard about, or read their name on publications - so that you can put a face, so to speak, to the name and to the science that you’ve known. And I’ve met some old friends, particularly from the group where I did my PhD. I avoid travelling not to abandon my family, but I do always find it surprisingly enriching whenever I do go somewhere. There have been a few other travels that I have done since then, mostly small and with a maximum one-night stay. First - this year’s edition of our Biotechnocentre meeting - the regular event about which I’ve written here already once, because I’m particularly fond of its mixed scientific-social format. And then, just last weeks, I went to two small French meetings, one in Strasbourg and one in Paris Saclay. It was snowing during the second meeting, the buildings on the other side of the large window of the lecture hall growing progressively white as the talks went on, the snow giving the occassion a somewhat festive feeling.
I have my habilitation defence next week - it makes me very scared, but, in some way, I’m looking forward to it, particularly to the social aspect of it - meeting people, talking, hopefully celebrating a little bit. I’m told it’s a formality, but I’d like to use it as an occassion to mark our move as a family to France and the first three years of living and working here.
I saw a little video the other day of an actor, I think it was Alan Rickman, the guy who played Snape, giving advice about how to be a better actor. What he said, or in any case what I remember from it, is that to get better at acting, you shouldn’t limit yourself to studying acting. You should go to museums and read books, and do other things that make you grow as a person. And then, out of that, the acting might get better as well. I found it wise, although there should probably be more emphasis not just on intellectual, but also on moral development. And, of course, I do think that this sort of advice applies to science as well. If we want our science to get better, what we need, I think; is not just studying science, but studying, reading, living, doing other worthwhile things that make us more of a person. And, out of that, the science might just get a little better as well.
Binding regions in ignored flexible protein tails: a case of XRCC4 and XLF (26/09/2024)
Just finished reading this paper, which I think is a very interesting case study in protein biochemistry. The authors looked at, previously largely ignored, flexible tails of two DNA-repair proteins, XRCC4 and XLF, identifying in these tails various specific binding regions. These regions, mapped at aa resolution, were shown to mediate either homotypic interactions, or heterotypic interactions (with DNA or with specific proteins that belong to the same DNA-repair complex). These interactions are transient/weak but could become significant, when the partners are brought together through more canonical structural interactions. In the end, this increases the number of possible binding events between the partners of this DNA-repair complex. As these events can happen within as well as between individual copies of the complex, they can facilitate clustering of multiple such complex copies together. This kind of protein condensation is in agreement with classical biochemistry: specific binding events, measurable affinities, avidity and cooperativity effects. As the core protein:protein interactions (those that are relying on highly conserved interfaces and motifs) are getting more predictable with computational approaches, I have a feeling we will have to focus more and more on such ancillary contacts, which might be more plastic in evolution and less predictable. NMR seems perfectly suited for this (and I’m saying this as someone who has never used NMR) - it would be nice to use some NMR of that kind for our projects! Are these ancillary contacts functionally important? The authors show that when you get rid of them, you do get a slight if measurable effect on DNA break repair in a cellular assay, and the effect is much greater if you additionally destabilise the complex. This points to these weaker interactions as increasing robustness of the system, which I can easily imagine is important, if not in an isolated cell, then at some point in the development of complex organisms, leading to the relative conservation of the identified interaction regions. Congratulations to the authors of this very interesting work, which was a pleasure to read as well.
Protein filamentation (23/07/2024)
It’s been already a week since our paper on filament formation by the BTB domains of some ZBTB-family proteins is out, but I’ve only just come back from holidays. For more details on our paper, see the summary that I’ve just posted in the “Publication summaries” section of my website. Here follow more general comments about protein filamentation.
When we hear of ‘filaments’, we typically think first of cytoskeletal filaments, which are abundant, long, and are (dis)assembled in a regulated manner. But many other proteins with a tendency to homomultimerise in one dimension are known. This is often called ‘polymerisation’. I prefer to speak of ‘filament formation’ or ‘filamentation’, because polymerisation implies a particular size range (poly as opposed to oligo), but what is really distinct about these assemblies is their shape and topology. ‘Dynamic filamentation’ refers to a particular subtype where filaments (usually of limited size) form and dissociate spontanously without additional factors. Such are the filaments formed by BTB domains that we characterised, such are the filaments previously reported for domains such as SAM or DIX.
Filament formation is ultimately a question of topology/geometry: if the homotypic interfaces are located on a protein in such a way that you can always add one more subunit without potential clashes, you have a potential filament. But whether such a filament will exceed a few subunits depends on affinity & local concentration. There are many geometries that could give rise to filaments. The best studied is ‘head-to-tail’ geometry: the front of a protein domain interacts with the back of another copy of the same domain. See this lovely review by Mariann Bienz, one of the pioneers of the field, on protein domains that make filaments with the head-to-tail topology. In the case of the BTB domains that we characterised, the topology is different: they first make head-to-head dimers (which, BTW, is the most easily evolving and abundant type of homooligomerisation - see my post about protein homodimerisation at the bottom of this page for details), and then the dimers connect with each other through tail-to-tail contacts.
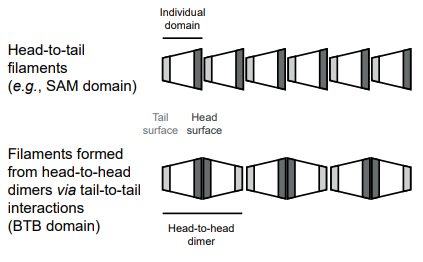
There are other topologies that give rise to filaments too. All this is important for their evolution. And also for prediction of filamentation with AlphaFold. AF2 or AF3 can work (doesn’t always do, but can), but has to be done in a way consistent with the expected topology. Head-to-tail polymers can be predicted by modelling a dimer: a protein might be a filament if you get a head-to-tail dimer with an angle allowing extrapolation to a filament rather than a ring. This is the clever approach taken by Emmanuel Levy’s lab and colleagues in the recent AlphaFold2 prediction-based study. For filaments built of dimers you need to model at least a tetramer. And so on. We comment on this in the discussion of our paper. Moreover, I think that in addition to 1-dimensional filaments, there might also be systems where proteins homomultimerise in 2 or 3 dimensions.
Is filamentation more common than we know? I think so. 1) They emerge easily in evolution (see this fascinating paper, again from Levy and colleagues). 2) They could explain some instances of foci in cells. 3) They might have been overlooked, because filament formation often causes insolubility on overexpression. This classic paper is a good example of the last point. The domain was insoluble, they fused a fluoresc. protein and did mutagenesis, found a solubile mutant. The mutant crystallised as a filament with the mutation at the interface, weakening the filament & making it soluble. Which, by the way, is also a good illustration of why crystallisation is a useful technique for filaments: crystal contacts might recapitulate real homotypic contacts, even if these are weakened by a solubilising mutation. But one has to be careful. Some crystal filaments might be a crystallisation-induced transformation of a ring-like multimer (see e.g. P65 or P61-symmetric crystal forms of ATPase domains). And being careful is generally a good idea when dealing with potential filaments. One has to think of artefacts of overexpression, etc.
A key technique for studying filaments is, of course, electron microscopy. As filaments are generally helical (you have to be helical or linear; otherwise, your multimer will circularise sooner or later), helical reconstruction of EM data comes in handy. SAXS and AFM are also useful in our hands.
One might add reason no 4 to why filaments could be more common than we think: because they are potentially functionally interesting. They concentrate a protein (in a specific manner!); they can give rise to avidity & cooperativity effects; they can create large structures with emergent physical properties.
Some general things we have noticed when working on filament-forming proteins (ZBTBs and others): 1) problems with solubility & aggregation, 2) predictability with AlphaFold2 or 3, but not always, 3) submicromolar or around micromolar affinity between protomers, 4) be careful with fusing not monomeric GFP as it amplifies foci (on the other hand, this might sometimes be useful for screening purposes).
CBM retreat, 16th-17th May 2024 (04/06/2024)
We had a two-day retreat of our institute, the Centre de Biophysique Moleculaire, held in a resort about 45 minutes away from Orléans, in the Sologne. Have you read the book Le Grand Meaulnes (translated several times into English, each time under a different title). Well, Le Grand Meaulnes is set in the Sologne, a region of forests and ponds south of Orléans. It was the first retreat since I joined (which was in 2021) and, in fact, the first, I’m told, that has been organised by the CBM in 9 years. We had scientific presentations by all the departments, some administrative stuff, long meals, a party. All in all, it was a lovely, joyful occassion. And the nicest thing, perhaps: a little surprise event for our long-time maintenance man, Justo, celebrating his many years at the CNRS with some videos and speeches sent by former colleagues. He, and two other long-time maintenance/administrative employees, Patricia and Martial, were awarded one of the medals of the CNRS for their contribution to the life of the institute over many years. A very moving occassion.
Alain Nicolas symposium, 11th March 2024 (09/04/2024)
I’ve been meaning to write something about a one-day symposium that I attended in Paris the other day, which celebrated the long career of Alain Nicolas, a French geneticist (who is still active and has recently moved from Institut Curie - where the event was held - to Niece). Now when I finally have a moment to write a few lines, unofrtunately, I don’t have my notebook with me, in which I jotted down some of my impressions. Anyway, I was quite impressed by this occassion and very happy to have decided to register for it in time, despite not knowing Alain Nicolas personally and not being from the field (genetics of recombination). It was a family event, in the sense of a scientific family: the speakers were recruited from among Nicolas’ coworkers (including his postdoc supervisor, the Noble-prizewinner Jack Szostak, who joined via zoom; several other American collaborators were there in person though), trainees, and colleagues. I could sense that we are celebrating someone special, a dynamic and inquisitive scientist, a good organiser, a faithful collaborator, one of the leaders of his field. Nicolas is a heir to a double heritage: the French one, that of the geneticist Jean-Luc Rossignol (Nicolas organised an event to commemorate him when he died in 2016), and the American one - that of Jack Szostak and a number of other figures whom Nicolas met during his postdoctoral stay in the Boston area. Someone mentioned that Rossignol, from whose lab in Orsay (near Paris) Nicolas originates, was, at the time, universally famous among geneticists, at least those working on recombination, and once he came for the first time to a conference in Cold Spring Harbor, everyone was intrigued, knowing him previously only from publications. He worked on the model fungus Ascobolus immersus, but Alain Nicolas moved to yeast and then I think also human cells in his later career. What a pity that Rossignol remains almost forgotten outside his direct circle now - it’s difficult to find anything about him on the internet.
In a way, this day was an exposure to the kind of science that is going away: science forged during conferences and through strong bonds between relatively small groups of scientists (some of them with imposing personalities) working on a given problem. Sure, conference still exists, but all the fields are much larger, the connections weaker. There were probably also negative sides to the way science used to be done compared to now, but this was the day to celebrate the good ones and learn from them. I took a lot of notes - scientific and also just ‘sociological’, if you like - and came back to Orleans very happy.
An additional reason for enjoying the symposium was that it was held in (or in any case next to) Institut de biologie physico-chimique (IBPC), an institution about which I have read a lot in the past. Founded in 1930 by Jean Perrin (a physicist and Nobel laureate) with the financial support of baron Edmond de Rotschild, it served a bit later as a model for the CNRS, the huge institution in which I happen to work, but is also interesting in itself, or rather through some of the personalities working there. I’m thinking especially of Louis Rapkine, an enigmatic, but apparently very amiable, researcher who sadly died young in 1948, and is known not so much for his research but for helping the exiled Jewish scientists, for his passion for culture (literature, music), left-wing, humanistic, but also somehow spiritual leanings. I remember reading a sort of obituary or memoir about him written by Leon Edel, the biographer of Henry James - I have to find it again. Outside the IBPC/university campus where the symposium was held, in Rue Pierre et Marie Curie, there were posters/panels about the history of IBPC, which I have now found online.
Filmmakers and scientists (29/02/2024)
I took a week of holidays to spend it with my family. During that time, I managed to read a little bit and finished a book I had started a long time ago, the biography of the French film director Éric Rohmer (1920-2010) by Antoine de Baecque and Noël Herpe. I don’t know how long it had taken me to read it, but probably several months. Mainly for my lack of time and the sheer length of the book (around 550 large pages), not for it being boring. Not at all! On the contrary, it was really enjoyable and made a nice break from science. I read it in the English translation published by Columbia Uni Press. I don’t know about you, but for me pastime reading is the one enclave of freedom, separate from work and any other duties. Having said that, I would say that filmmaking, at least of the type that Rohmer practised, is not that far from science; and neither is, in fact, any good art - a thought, perhaps naïve, to which I will return at the end of this blog.

My reading Rohmer’s biography has gone along with watching a few of his films (found in our local library) every now and then over the last several months. We have a few more that we would like to watch. I guess my main motivation has been that, now living in France for the first time, I feel like I should learn a bit more about the French culture, including the 20th century, of which my knowledge is rather patchy (I like some writers and some philosophers, but I don’t know much about more popular culture). Films from the French new wave felt like a place to start, and of the directors I have read about, Rohmer felt like a kind of cinema I would like. And I did like his films, a lot. My wife, for the record, much less, and it’s a testament to her love that she watched them with me anyway. She found the films a bit imperfect, amateurish. For my part, I like the way de Baecque and Herpe put it in Rohmer’s biography: “Rohmer preferred the actor’s irregularities that gave his story the imperfection of life”. “The imperfection of life” - that’s it!
It’s hard to describe Rohmer’s films to those who haven’t watched them. People talk a lot in his films, and live through small dilemmas of everyday life, often connected to relationships. Rohmer is someone who likes people, is curious about people, about how they behave and react, and he just shows that, in a fairly simple but somehow captivating way. My favourite of his films so far has been The Green Ray (released one year before my birth) - a purposefully banal story that nonetheless makes you think (about the interplay of chance and purpose, for example) - and smile.
A strange question perhaps, but one that motivated me to write this blog: is there something that an aspiring scientist like myself, a young scientist who might fail any moment but otherwise enjoys science, can learn from Éric Rohmer in terms of how to do science? I think there is, based on an analogy between Rohmer’s filmmaker’s practice and career and that of a scientist’s. Films, like science, are made in teams, for example, and Rohmer seems to have been someone who was good at bringing people together through rituals (5 pm tea breaks), through discussion on any topic whatsoever, through friendship. Also, one needs to get funding to make films, just like one does to do science, and getting funding is fuelled by recognition. Most films that Rohmer made were done quite cheaply. Being economical and careful gave him freedom, because it was easier to collect modest amounts of money, and the pressure for returns was smaller. In terms of recognition, Rohmer generally kept doing what he thought was good cinema and didn’t seem to try to please others. He was sometimes ridiculed and often misunderstood. Perhaps he didn’t take feedback sufficiently into account, but there was also virtue in his faithfulness to his own vision and values. He did make some films that have been judged as failures. But also many that have been acclaimed by critics and loved by cinema-goers. It’s a lesson in not caring about recognition but just trying to do good art. Or science, for that matter. If needed with modest means, but strong values and a sense of purpose.
When reading the biography and watching Rohmer’s films, I have been struck by these similarities between a filmmaker’s and a scientist’s fate. But I also kept thinking about a more general unity of science and art. Isn’t filmmaking and perhaps most other artistic activities about capturing, naming, and/or showing something of reality that has not been captured, named, shown before? That seems to me to be on the same continuum as science. Moreover, quite often – as in the case of Rohmer’s historical films, for example - it takes a lot of actual background research (of historical but also technical nature) to do art, in addition to artistic sensitivity and skill. Now, I like a lot a philosophical approach called virtue epistemology, which teaches that gaining knowledge and discovering truth requires certain virtues and skills, some of more moral kind, like selflessness or humility, other more “technical”, like perceptiveness.

In a volume of essays from this genre entitled “Virtue Epistemology Naturalized: Bridges Between Virtue Epistemology and Philosophy of Science” (Springer 2014), there is one by Shannon Vallor, who speaks of “perceptual responsiveness” as the basis of good scientific observation. Vallor, a philosopher from Edinburgh, gives the famous 20th century geneticist, Barbara McClintock, as an example (another is a pioneer microscopist from the 17th century, Robert Hooke). This is the description of scientific practice that Barbara McClintock gives: “You are so absorbed that even small things get big… Nothing else matters. You’re noticing more and more things that most people couldn’t see because they didn’t intently go over each part, slowly but with great intensity. It’s the intensity of your absorption.” And elsewhere, complaining about the attitude of some scientists: “I feel that so much of the work is done because one wants to impose an answer… They have the answer ready, and they want the material to tell them”. Anything else it tells them, “they don’t really recognise as there… If you’d only just let the material tell you”. These are all words by the geneticist, Barbara McClintock, who, through this kind of openness to reality and resistence to preconceived ideas (but also through her technical skill, familiarity with her specific material, i.e. maize, her mathematical analysis, her sheer hard work) could provide a description of the biological process of gene transposition. But these words could, just as well, have been written by a writer. Or a filmmaker. Indeed, to me, this kind of openness to reality is what brings scientists and artists together, definitely artists like Rohmer. Of course, the aspects of reality that artists are usually interested in are different from those that interest most biologists. Artists, writers, filmmakers are more interested in human relationships, in feelings, in our experience of the world and ourselves, in things like chance, fate, and providence, in what it means “to be rather than not to be”, in the moral complexity of human actions, etc. Psychological, metaphysical, moral questions. But the basic tasks of seeing, naming, showing the multi-layered and multi-dimensional reality in which we live are not that different. “It’s not myself I see in my films, it’s the world I’ve filmed”, said Éric Rohmer.
Inhibition of SUMO1 by its own N terminus (26/02/2024)
We started a journal club a few months ago. One meeting a month. In the 3rd session, we discussed the recent pre-print about the role of the flexible N-terminal tail of SUMO1 in regulating SUMO:SIM interactions. It’s a very nice combination of biochemistry, NMR, and simulations (and a little bit of in vivo). The prepring is here. There are two main molecular differences between ubiquitin and its homologue SUMO: 1) SUMO behaves like a beta-sheet that misses a beta-strand and it can interact with SUMO-interacting motifs (SIMs) that bind in a beta-strand conformation; 2) SUMO has a flexible N-term tail.
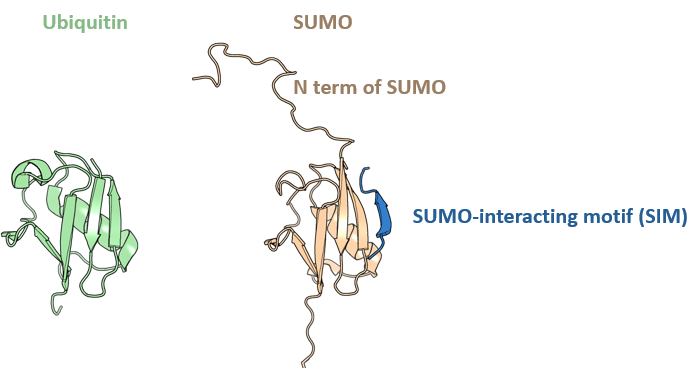
I should add that in humans there are 3 main paralogues of SUMO, with SUMO1 being a bit different from SUMO2/SUMO3. All bind to SIMs. The basic idea of this study is that the N-terminal tail of SUMO1 (but not SUMO2/3) inhibits the binding of SIMs to SUMO, providing regulation.

The inhibition seems to be due to a transient, ionic intramolecular interaction between the N-terminal tail and the area around the SIM-binding grove. This is nicely suggested through NMR, mutagenesis, and molecular dynamics. This is conserved in the sole S. cerevisiae SUMO Smt3 and in the C. elegans SUMO SMO-1. It doesn’t extend to the human SUMO2, but the authors show that, probably, if the SUMO2’s tail gets phosphorylated, it will get the right electrostatics to start inhibiting SIM interactions. It was my turn to present this time. Since one of the last authors of the paper is Frauke Melchior, one of the discoverers of SUMO, I made a short historical intro including this nice interview. I recommend the paper & congratulate the authors (and thank our journal club attendees).
Being a young scientist (26/02/2024)
I’ve read two interesting advice pieces written by established scientists, How to be a good Principal Investigator by Lawrence Banks and Things you should learn in graduate school by Ramanujan S. Hegde. I’ve been once in a (purely scientific) talk by Hegde and was very impressed; I’ve never heard or seen Banks, but I know of his work in virology. They belong to different generations and one can sense it a bit in the two texts, but both articles are interesting and useful. I’ll write a few things down here, not to forget them.
Banks’ article is meant specifically for group leaders. He divides his advice into three parts, tailoring it to new PIs, mid-career PIs, and late-career PIs. The advice is very different depending on the career stage, but there are some common threads. Overall, there is an emphasis on providing motivation to the team, on choosing the team well (so that older students are role-models to the younger one), and on leading by example (which, for new and ideally also late-career PIs, includes keeping working at the bench). Team building through regular coffee breaks or after-work get-togethers comes through as more important than regular group meetings, which Banks doen’t particularly value. It’s more fruitful, he says, to create an atmosphere in which people are eager to share and discuss with others their results as these are “hot of the press”. “Foster an environment - Banks advises - where lab members are comfortable coming back from developing a western blot and sharing their results: ‘wow guys, look at this! - this is the second time I’ve done this and the result is exactly the same - this is completely unexpected and something really new - what do you think?’”. One way to ensure such an environment is not to let the lab grow too big, beyond what is managable; and to always have time for your people, even when you are a “late-career PI” busy with departmental duties etc. Banks emphasises a few times that being a successful scientist involves working very hard, also at weekends, and that students should ideally come to understand that by themselves. This was where I wasn’t sure if I agree. Working hard, yes, but perhaps within reasonable limits. I don’t know if I’m a successful scientist, but I’ve almost never worked at weekends in my life, and I’m grateful to my mentors for letting me not do it. But I understand that Banks’ experience is probably different.
Now Hegde’s article, which is technically meant for PhD students, but I think it’s a very valuable reading to any aspiring scientist. His main message is that more important than mastering any particular technique is acquiring a “scientific instinct”: a skill that includes finding problems that are important, interesting, and doable within your constraints; then dividing the problems into short-term projects; then knowing how to learn the techniques that one needs to execute the project (the answer is: learn enough about the technique in question to be able to customise it to your specific question); and, finally, presenting the results in simple, logical, compelling prose. Because scientists, to stay productive and successful, are obliged to often switch projects or even fields and adapt to constantly changing techniques, Hegde’s emphasis is on what is enduring and transferable. A PhD student usually begins with a project allocated to them by an advisor but should ideally move to a position when she or he is able to propose important and feasible project by her/himself, and not only in your direct area of research but also more broadly. Hegde argues that the “scientific instinct” is not some irrational intuition but the ability to act in a reasoned and logical manner in the world that is full of uncertainties and where information is always incomplete. Thus what seems instinctive in some exceptional scientists is probably highly logical and reasoned, it’s just that the rationale remains unarticulated. And you can learn from such exceptional scientists, perhaps by encouraging them to articulate how they reached that particular decision. Generally, we should - Hegde says - learn from others, not just our direct supervisors, but any people in whom we notice qualities that we would like to develop ourselves. In addition to mentors, we can also learn by examining how some specific decisions have been reached in the past - those leading to failures and success. One interesting point that Hegde makes is that successful scientists often are skilled in making inferences from analogous problems in other areas - which means you have to read widely also outside your domain. And finally: when writing an article, you must put yourselves in the shoes of someone who doesn’t know your reasearch, and you must adapt your narrative (composed of simple, logically connected paragraphs, each containing a single main point) in such a way as to logically argue how your results support a given hypothesis. Since it has to be a consistent narrative, Hegde doesn’t see a point in starting with describing results or making figures first; rather, he says, write in the same order as the one in which the text is read, from the title to the discussion, adding and adapting figures to the linear narrative.
I think these two texts, very different and aimed at different audiences, and written by authors from different generations and with different personal experience are actually quite complementary, one emphasising mainly the human and social aspects of a good environment, another - the individual skills one needs to develop. I’ve found both texts really illuminating. Of course, in the end, science is not only about science, and not only about success (indeed, being a good scientist, or at least a decent human being who also does science, often means one needs to fail). Science must be reconciled with other important areas and other domains of human and social life, particularly with the world of values, justice, kindness, morals.
Modelling oligomerisation with AlphaFold (19/02/2024)
Paper from Emmanuel D. Levy and his group are always very interesting. This study, now published in its final version, is about modelling protein oligomers in a high-throughput manner using AlphaFold2. It’s worth a read, not only because of the excel file with predicted oligomerisation symmetries for most human proteins and those from three other species (E. coli, P. furiosus, S. cerevisiae). The approach the authors took to predict oligomeric states of proteins with AlphaFold2 with reasonable computational resources was to first model dimers, analyse the confidence scores, and - if those were sufficiently high - analyse symmetry of the dimers. Dimers with head-to-head (C2) symmetry were identified as potentially just dimers, while dimers with head-to-tail symmetry were identify as potentially ring- or filament-forming. By extrapolating the head-to-tail dimeric models, possible numbers of protomers in a ring were proposed (trimer, hexamer, decamer, etc.), or - if the oligomer didn’t close into a ring - a protein was suggested to form a filament. Interestingly, a standard version of AF2 - trained on single protein chains - was used instead of the ‘multimer’ one, as the gain with the latter was apparently limited and to avoid the situation where the training PDB set contains quarternary structures that the study predicts. Like always in the Levy lab papers there are many interesting general observations, like e.g. that the number of possible tertiary folds seems much more limited than the number of possible quaternary arrangements - AF2 identifies many new quaternary but few new tertiary structures.
One should note that not all possible oligomeric architectures are covered by the adopted approach: higher-order arrangements built of symmetric C2 dimers would not be predicted, for example.
All in all, it’s a very interesting paper with a lot of new information.
PARP1 condensates (12/02/2024)
PARP1 is the DNA-damage sensor that binds to DNA breaks and synthesises protein modification made from long poly(ADP-ribose) (PAR) chains. It is also my friend since my second postdoc, when I worked on PARP1 and its interactor HPF1 - a very interesting protein, by the way, which forms a composite active site together with PARP1 and changes the way it makes PAR chains.
I try to follow the literature on PARP1, especially as we are trying to do some work related to it in my current group as well. Among the different papers I have read on PARP1 lately, the recent one in Cell from Simon Alberti’s group has been the most interesting. Here is the link. Really, a very interesting paper with elegant biochemistry. Upon binding to exposed dsDNA ends mimicking a double-stranded DNA break, PARP1 can multimerise at relatively low concentration into large isotropic complexes that manifest as condensates observable under microscopes. The PARP1 multimers formed on dsDNA binding are mediated via specific interactions between structured domains from different PARP1 molecules. Some of these domain:domain interactions had been proposed before, occurring in cis within the same PARP1 molecule, but it looks like they could occur in trans between different PARP1 molecules, thus non-covalently cross-linking multiple PARP1s into larger objects, beginning with a dimer. There might be further unknown interfaces needed for this process (and perhaps some are hidden as crystal contacts in existing crystal structures?). PARP1 is a large multi-domain protein. It’s interesting that each of the domains except for the catalytic domain is needed for the condensation. In addition to a dsDNA that mimics double-strand breaks, condensation of PARP1 was also observed with nicked DNA, resembling single-strand breaks. What is the functional importance of this phenomenon? Nice optical tweezer-based experiments suggest that a PARP1 condensate prevents two sides of a double-strand DNA break from separating, plus the condensate could provide the initial repair environment. In cellular experiments, PARP1 seems to share with KU70 the role of preventing DNA ends from separating after a cut, the double loss of both proteins leading to the loss of bridging. PARP1 is also an enzyme, which gets activated upon DNA break binding and generates poly(ADP-ribose) (PAR) chains from NAD+. It looks like PARP1 is catylytically active in condensates and PAR chains can weaken or altogether prevent condensation. But then there are proteins, like FUS, that can come in to stabilise the condensates by interacting with PAR chains, and one can imagine a complex evolution of the initial condensate in cell on arrival of different proteins. Trapped PARP1 would be PARP1 in condensates that cannot mature beyond the initial stage. Overall, I really enjoyed it & it made me ask myself a lot of questions when reading. Protein condensation, which may or may not involve phase separation, has always seemed to me a somewhat esoteric phenomenon, and I know that it is controversial. But condensation mediated by specific domain:domain interactions is easy for a structural biologist to swallow, and it makes sense that multivalent molecules should be able to crosslink (or ‘percolate’) into large isotropic networks that manifest as spheres in vitro or foci in cells. Congratulations to Nagaraja Chappidi, Simon Alberti, and all other Authors.
Modelling domain:peptide interactions with AlphaFold2 (24/01/2024)
I like to think that, as a rule, at least one partner involved in a binary protein:protein interaction has to use a folded domain. There might be exceptions to this rule - biologically significant interactions between two regions that cannot be characterised as a folded domain - but I think they would be rare or perhaps not rare but difficult to detect and characterise with the tools that we use as they would probably be weak and serve only auxiliary roles in a system already stabilised by more canonical interaction. The reason I think that in more canonical interactions at least one of the partners tends to use a folded domain is that a domain can have a binding pocket with which it can at least partially surround a smaller ligand (such as a short linear protein motif) to achieve sufficient interaction surface and/or sufficient number of individual noncovalent bonds to get a meaningful interaction, which would be difficult if both partners were small. Now regarding the other partner, it might also be a folded domain or it could be just a short linear motif, typically embedded in a disordered region, which becomes more rigid upon binding to a domain. These kind of interactions, which are usually weak/moderate thermodynamically (in a micromolar range) and transient kinetically, could be described as domain:motif interactions. We know very many examples of such interactions and it’s possible that they are the most abundant type of binary protein:protein interactions, at least in higher organisms with complex interaction networks. In the SUMOylation system - by which I mean enzymes, substrates, and readers associated with the protein post-translational modification (PTM) called SUMOylation - interactions of this type are key and they include interactions between SUMO and the so-called SUMO-interacting motifs (which can be found in SUMO reader proteins and in SUMO E3 ligases), but also interactions between the SUMOylating E2 enzyme UBC9 and the so-called SUMOylation consensus motif on substrates. With the recent advances in AlphaFold2, many people have been asking if this tool can be used for predicting protein:protein interactions, including domain:motif type, and we also try using it to study these type of interactions in the SUMOylation system. Not being a computational biologist by training, I try to follow the literature by experts in the field, which by the way is moving fast. Recently there have been two papers out - both previously released as preprints - that deal specifically with the question of domain:motif interactions: this one and this one. They are partially overlapping but mostly complementary and I think it’s great to read both. I’ve finished the first and am halfway through the second. The first is by Hélène Bret as the first author and Jessica Andreani and Raphaël Guerois as supervisors, working not far from us in Gif-sur-Yvette near Paris. What is great in the paper is not just the results but also a very useful introduction and similarly useful discussion, in which the recent literature and the available methods and tricks are discussed in an accessible way. Indeed, AlphaFold2 seems to be an “art” as much as “science” and a lot is about finding the right protocols to run it most efficiently. I noted down a few papers with which I have to catch up. The bulk of the results presented in the paper is based on a set of 42 interactions between a structured domain and a short motif embedded in a disordered region. This set comprises new structures not included in the training set of AlphaFold2-Multimer and also lacking homologues in that set. This set is then used to evaluate how good AlphaFold2-Multimer, applied in different ways (with full-length proteins or fragments, with or without multiple-sequence alignment, with different types of alignment, etc.) is at predicting such interactions. One interesting conclusion - which aligns with our experience and probably those of some others, but we haven’t analysed it systematically yet - is that prediction works better if sequences are limited to relatively short fragments spanning the interaction regions (ideally not more than 100 aa on the motif side; for the domain, one can, and probably usually should, take the whole delimited domain). One can scan the disordered sequence with such windows to find the motif if its location is unknown. A multiple-sequence alignment (MSA) of the disordered fragment is particularly beneficial if long sequences are used, but if sequences are delimited, it makes surprisingly little difference and just having the MSA of the domain is enough. (Sometimes including an MSA of the disordered fragment might even be detrimental, as when it makes AlphaFold2 fold the interacting motif in a structure that is not compatible with binding! - there are some interesting examples of that discussed in the paper). In the words of the authors, “Such a performance suggests that the structural and evolutionary properties of the receptor match well with the peptide sequence irrespective of its conservation pattern”. As far as I understand, there is a lot of discussion in the field which metrics it is best to use to rank AlphaFold2 models and identify potentially real ones. In this paper, they use a linear combination of 0.8 * ipTM + 0.2 * pTM, which seems to work well, so it might be a useful, simple metric to use. Thanks to the authors of these lovely papers!
Review article, article review (21/12/2023)
I’ve finished revising and re-submitted a general review on chemistry, mechanisms, and evolution of protein post-translational modifications (PTMs). This brought back the memories from 12 years ago or so, when I had just finished my undergraduate studies and did a 6-month internship at the Weizmann Institute in Israel, in the group of Joel Sussman.
It was a great, formative time for me. I shadowed a PhD student and did some enzyme kinetics measurements and crystallisation with him, learned to write scripts in Perl, and - which is why I started thinking about it again - wrote a review article - the very first scientific article of any type in my life. The topic was proteolytic stability of intrinsically disordered proteins in vitro and in cells. It started with Joel asking me for my opinion on a paper he had to review. Nobody had ever given me such a responsible task, so I approached it (too) ambitiously, read widely around, and wrote a long (too long) opinion on the article. Joel suggested we transform it from a review of a paper into a review article on the topic that the paper was about. I had then recently learned how to make simple schematics in a freeware equivalent of Illustrator called Inkscape, so I tried to make use of it for illustrating my little review. I don’t know if what we wrote there still remains up to date after 12 years, but I have fond memories of writing it, discussing it with supervisors and getting their feedback, and then of seeing it published. Since then, I have participating in writing a few more review articles, but the one now is special - and closest to the one 12 years ago - in that I did it mostly by myself and put extra effort. Let’s see what comes out of it. PTMs have been an interest of mine for many years, the main unifying theme that connects different projects I have been doing over the last 12 years or so, since my undergraduate studies and that internship with Joel. Studying different particular PTMs, first in bacteria (protein arginine phosphorylation) and then in eukaryotes (ADP-ribosylation, ubiquitylation, SUMOylation…), I started asking myself questions about the chemical nature of these processes, and how it is connected to, on the one hand, their catalysis, and, on the other hand, their emergence and development during evolution. In short, what I think is interesting is that most major PTMs are chemically analogous in that they all involve some kind of nucleophilic substitution reaction where a protein reacts with a donor molecule present in the cell (usually present there for other reasons, and present already before PTM reactions emerged - examples of such donors include ATP, NAD, SAM, acyl coenzymes A, UDP-sugars, etc., all of which are involved in metabolic processes in addition to protein modification). In these donor molecules, one can distinguish a good leaving group, which leaves during the reaction, the rest of the donor ending up covalently attaching to the protein. So enzymes that catalyse PTMs tend to perform similar tasks, binding the donor in such a way that it promotes the departure of the leaving group and the nucleophilic attack, and often they deprotonate an amino-acid residue on the protein to make it more nucleophilic and attack the donor more efficiently. In the review, I try describing these chemical and catalytic analogies, but also analogies between different PTM systems in terms of how the substrates are recognised by the writer enzymes, how the modification might be reversed by eraser enzymes, and how the product is recognised by reader proteins. And, lastly, I try to discuss the evolutionary emergence of these processes, where one part of the story is that the suitable “activated” donor molecules had existed in primordial prokaryotes to serve in primary metabolism and then became repurposed as co-substrates of enzymatic protein modifications. That’s a very brief summary of the review, which will perhaps be out in a few months. There will be some figures, too. Now I have a license for the real Adobe Illustrator, but I still make the same sort of simple schematics to illustrate the processes I write about.
So that’s one thing I’ve been doing in the last few days. I also had to do some things in the wet lab (mutagenesis and some recombinant protein production, autoclaving media, etc.), and today I wrote a review of an article. Since very recently, I am (very sporadically) asked to do it directly by journals, which is flattering but also some responsibility. With the review article and the article review now submitted and this blog post finished, tomorrow I start two weeks of holidays for Christmas.
Paper on PARP4 (01/12/2023)
I’ve finally read this study of PARP4 by Léonie Frigon and John Pascal and strongly recommend it to those interested in PARPs and ADP-ribosylation (one of my interests, especially during my ‘previous life’ as a postdoc). PARP4 is an odd cousin of the canonical PARP protein, PARP1, sharing a larger homology region with it than any other of 17 or so PARP family members in humans. At the same time, PARP4 is quite diverged from PARP1 in terms of cellular location and function. Previous work, largely from the Pascal lab, showed that PARP1 is an amazing allosteric machine that is very tightly regulated. Now PARP4 can be seen as an example of evolution duplicating and rewiring this molecular machine, keeping some functions and chaning or modulating others.
I enjoyed the thoughtfully crafted and cautious text that leaves room for discussion and interpretation of results, going through various important aspects of PARP4 and how it seems to differ from PARP1, including the regulation by HD, the absence of the PAR chain extension function in PARP4, the potential roles of WGR and BRCT domains. For example, it looks like the HD of PARP4 does not inhibit its catalytic activity much (unlike in PARP1), and the WGR apparently lost its nucleic acid-binding function, while BRCT retains it, possibly prefering vault RNA over other nucleic acids, although that is not completely clear.
Earlier this year we published a paper that analyses AlphaFold2 models of all PARP family members, including PARP4. I’m sorry if it has in some ways impinged on the apparent novelty of this article by Frigon and Pascal or caused any stress with their publication. I don’t think it did impinge on the novelty. Indeed, we only now get a real experimental analysis of the N-terminal catalytic region of PARP4, combining crystallography with functional in-vitro assays.
Working at the bench (27/11/2023)
I do hope I will be able to continue working with my own hands at the bench still for some time, rather than focussing only on administrative and supervision roles. I do enjoy the wet lab a lot and also think working with your own hands is useful for thinking about projects. There are many great group leaders who do not work at the bench any more themselves, but I would like to do it if I can. I’ve been in a meeting with a Nobel prizewinner for ubiquitin discovery, Avram Hershko, once, and he said that even now, after the big prize etc., and in his wisdom years, he works at the bench, usually from the early morning until around midday, where he begins to take care of other things. That sounded great. On this topic, a quotation I’ve found, about the pioneer of fly genetics, Thomas Hunt Morgan: “Throughout their careers Morgan and students worked at the bench. The investigator must be on top of the research if he or she is to recognize unexpected findings when they occur.”
At home and away (27/11/2023)
I do enjoy travelling, although much less since I have a family, unless, of course, we can go together. The last two months or so have been a period of several work-related trips, mostly short and nearby (all by train), but still amounting to quite some time away from home and the lab, and only one of the trips was together with my family.
One, alone, was to a local conference organised mainly by and for scientists from Tours and Orléans, the yearly Colloque Biotechnocentre. It was its 35th edition already. The conference is held in French and takes place on two consecutive days, with an evening (with a party) in between. There are poster sessions for PhD students and talks by invited guests from outside the region and local scientists, both more senior and PhDs. I had a short talk entitled ‘Why we study protein SUMOylation?’, or rather ‘Pourquoi étudier la SUMOylation des protéines ?’, because it was the first proper scientific talk that I’ve given in French, with much fear and trembling. But it was a nice experience in the end, having managed to do it and seeing that I have been mostly understandable. I enjoyed this particular yearly conference very much. I like its broad scope in terms of topics but at the same time the regional focus, and I like the multi-generational make-up, with both older (some retired for some time) and young scientists, and last but not least the fact that it combines an intensive scientific programme with a party and long French-style meals etc. At the same time, it is relatively modest in terms of venue etc. and not too expensive. I was very impressed by PhD students - their talks and posters were very well prepared and well presented and the atmosphere was friendly. By the way, I had a very similar impression at a PhD defence I attended in Rennes as a member of a jury in early November - of being impressed by the PhD students, both the one defending (she did it great), but also those attending the small party afterwards, being there for her.
Another recent occassion was a meeting of prize-winners of the French ATIP-Avenir programme. A great meeting as well, and what I particularly liked this time was that they invited some older scientists (some of them themselves ATIP-Avenir laureates from the past) to give talks that were not pure scientific talks but rather attempts to share experience of running a group and developing your research after the first grant. Two of the speakers whose names stayed in my head were François Parcy and Jean-Louis Mandel, but there were many others. There were also short talks by the ATIP-Avenir awardees themselves, including myself, presenting the selected projects. I should say that because of the rules of the ATIP/Avenir programme, getting an ERC grant means you don’t get any money from ATIP - but you still get a ‘label’ of an ATIP-Avenir programme and get to attend the events like this, which is nice (there were several people in this situation).
Finally, the last two trips - 5 days in Marseille on the biannual French structural biology meeting called BSI, and then a seminar in Basel, at the FMI institute, last Friday. But let me write more about the BSI conference. It was the second BSI meeting I have attended in my life, after the one two years ago in Saclay, which was one of the first things I did after starting the job in France. The BSI meetings are really good. Quite diverse in terms of biological questions and techniques (ranging from EM and crystallography to more niche biophysical techniques such as AFM or EPR), with a diverse set of speakers at different career stages (most of them from France, but there are always some invited speakers from other countries). It is a good opportunity to get a sense of how the field is developing, at least in France (e.g. this year - the growing interest in EM tomography and, obviously, in AlphaFold). I had a talk in which I presented, for the first time, a new topic in the lab. The talk went ok, I think, but I didn’t have it finished before the conference so the evenings before my talk were stressful and busy with rehearsals. There was a lovely ‘gala dinner’ during the conference in a museum, on the last floor, overlooking the harbour of Marseille.
It was at the BSI in Marseille that I had a conversation with one young scientist about science as a social phenomenon. This social dimension of science - the invisible hierarchies and codes that govern it - has always fascinated me. Conferences or visits at other labs are great opportunities not only to learn about new discoveries or establish collaborations - although they are both - but also to look at science for a moment with an outsider’s eye, which allows noticing various peculiarities of how sience works.
But there is also a price to travelling, especially being separated from one’s family and putting an unequal burden on the spouse/partner. I hope I will travel less in the future.
Nobels 2023 (06/10/2023)
Like every year, I’d been eagerly awaiting the announcement of Nobel prizes. Like everybody else, I was very happy to hear that Katalin Karikó and Drew Weissman were awarded, especially Karikó, but what I think made me even happier was the Nobel prize in literature awarded to Jon Fosse, a Norwegian writer whose thick Sepatalogy I began reading some time ago and which has enchanted me in a special way. A running joke is that Fosse’s new novel is easy to read, because you just need to read one sentence (there are no full stops in the whole long book). Well, easy it is not, but it is a particularly convincing example of a stream-of-consciousness narrative, and the writing has a singular trance-inducing rythm.
Grenoble revisited (04/10/2023)
We’re going to Grenoble today, just me and a colleague, for a couple of days, to do some SAXS experiments at the synchrotron. I think it’s been about 12 years since I first visited the ESRF synchrotron. It was before my PhD, during an internship I did in the group of Joel L. Sussman at the Weizmann Institute in Israel. We spent a few days in Grenoble, collecting data for two days or so and also doing a little bit of sight-seeing (we took a cable car to a base beneath Mont Blanc and stopped in Chambéry). I remember the Hebrew word “maruah” that we used when poorly diffracting crystals gave a smeary pattern. In addition to it being the first time at a synchrotron, it was one of the first encounters with France. Eating croissants in the ESRF guesthouse for breakfast, little did I know that I would one day live and work in France. I’ve been to Grenoble a few times since, but not in the last 5.5 years or so, so I’m happy to go back. And it’s the first time I’ll do SAXS there.
Added on 6th October: So now we’re actually in Grenoble, and almost finished and leaving very soon. It’s been a lovely couple of days. We’ve met some kind people at the beamline and outside of it, including some old friends and a postdoc behind the data of one of my favourite talks in the Aviesan meeting mentioned below. The scenery of the Polygone Scientifique of Grenoble, with mountains all around, and the atmosphere - all the scientists, including many young people, working here or just coming to collect data - it’s all very special.
Aviesan ‘Membrane-less organelles and liquid condensates’ symposium, Paris 25th September 2023 (26/09/2023)
I very much enjoyed yesterday’s one-day symposium on membrane-less organelles organised by Aviesan, one of those enigmatic French organisations that I haven’t figured out yet. What I’ve figured out so far is that it is an alliance of scientists that is reputed for organising very nice free symposia on different topics, and this was one of these. The main organisers of this specific symposium were Carine Giovannangeli, Yves Gaudin, and Dominique Weil. The symposium was held in Jacques Monod Institut in Paris (my first time there), so very conveniently near the Austerlitz Station where trains from Orléans arrive. I don’t know the field of biomolecular condensates very well and have been at times sceptical of some very general claims that I heard, but the symposium made me realise that there is a lot of nuance in the field, like, for example, an attempt to distinguish polymer polymer phase separation or mere binding-driven phenomena from (perhaps rarer?) liquid-liquid phase separation, or the realisation that ‘membrane-less organeless’ often result at least in part from defined and specific (even if weak) binding events, or the idea that there is sometimes some degree of order within these apparently liquid structures. So there is nuance and more theoretical and technical innovation, but there is also certain dynamism and audacity characteristic of new fields and new paradigms, and I enjoyed that aspect a lot. I think my favourite talks were those by Chloe Zubieta, Ariel Lindner, Fabian Erdel, Martin Blackledge, but I liked all of the others too, including by young scientists, which did a very good job (a postdoc Damien Glon, for example, and Anastasiia Skolbelikna). Overall, a great line-up of talks spanning a bit the whole field in France, with both some finished studies and some work in progress. The main keynote lecture by Anthony Hyman - the only speaker outside of France - was on Zoom but the technology worked well. I’m looking forward to more events like this.
Protein homodimerisation (20/09/2023)
A lot of proteins we see in nature are homodimers in which the dimerisation interface is formed by two identical surfaces, one on each subunit, coming together. This is how many proteins have evolved to be, and various functional advantages of homodimerisation (things like cooperativity, or higher stability, or easier folding, etc.) might have driven this evolution. But, generally speaking, for a feature to be selected for and optimised over time, it has to already exist in a primordial system, and perhaps if there is a bias towards that feature already at the start, it can be more easily picked up and optimised over time. Is there, therefore, an inherent bias for a random protein surface to associate with itself rather than with a different protein surface? This is a question posed in this somewhat old paper from Lukatsky et al. (the Shakhnovich lab). The paper contains a simplified model of a protein surface through which this question is investigated computationally (giving an affirmative answer), but there is also a common-sense argument, which I like, and which I think can be rephrased in the following way. What is special about a typical homodimerisation interface? That it has what crystallographers call 2-fold rotational symmetry (C2 symmetry), which means that all (or most) interactions are repeated twice. If residue X from subunit 1 interacts with residue Y from subunit 2, then residue Y from subunit 1 interacts with residue X from subunit 2. This repetition has implications for the likelihood of making homodimers, because, if you make certain favourable interactions by chance, they will be repeated twice. I think this is similar to what Lukatsky et al. say here: “it is more probable to symmetrically match a half of a random pattern with itself than with a different random pattern, which requires a full match”. If we imagine that dimerisation is pattern-matching, it is more probable to match half of a pattern (& symmetry does the other half) than to match a full pattern (which is what has to happen during hetero-dimerisation)
I think that the repeptition idea also implies that the rate with which an interface becomes optimised over time is higher for a homodimer than a heterodimer - because a favourable mutation that strengthens the interface would have an amplified effect (would effectively be a double mutation). As Monod and his colleagues working on haemoglobin already observed, symmetry amplifies effects. Perhaps this is the common-sense rationale behind another modelling study, this time from G. E. Schulz and with a more sophisticated model tracking evolution over time. This second study concluded that a simulated evolutionary process that leads to homodimerisation has the probability that is by a factor of 100 or above higher than the probability of an analogous simulated process that leads to heterodimerisation.